Журнал «Здоровье ребенка» Том 19, №3, 2024
Вернуться к номеру
Генетична схильність до метаболічно асоційованої жирової хвороби печінки
Авторы: Абатуров О.Є., Нікуліна А.О.
Дніпровський державний медичний університет, м. Дніпро, Україна
Рубрики: Педиатрия/Неонатология
Разделы: Справочник специалиста
Версия для печати
Літературний огляд присвячений висвітленню питання щодо генетичних факторів ризику, що мають зв’язок з розвитком метаболічно асоційованої жирової хвороби печінки (МАЖХП). Генетичні обстеження людини виявили 132 гени, серед яких 32 локуси міцно пов’язані з патогенезом МАЖХП. Встановлено, що ризик розвитку МАЖХП несуть однонуклеотидні варіанти різних генів, продукти яких беруть участь у ліпідному, вуглеводному обміні, підтриманні окиснювально-відновного стану, розвитку запалення й фіброзу тканини печінки, тобто є компонентами МАЖХП-реактому. Автори навели детальний перелік генетичних факторів, окремо виділивши серед них ті, що впливають на ризик виникнення МАЖХП та безпосередньо метаболічно асоційованого стеатогепатиту й фіброзу печінки. Автори акцентували, що саме однонуклеотидні варіанти генів протеїну 3, що містить пататин-подібний фосфоліпазний домен, трансмембранного протеїну 6 — члена суперродини 2 і 17b-гідроксистероїд-дегідрогенази 13-го типу характеризуються найбільш високим ступенем асоціації з МАЖХП (ВР > 1,6) порівняно з однонуклеотидними варіантами інших генів, ідентифікованих за допомогою досліджень генних асоціацій. Поєднання декількох поліморфізмів збільшує ризик розвитку і тяжкого перебігу МАЖХП. Адитивний стеатогенний ефект однонуклеотидних варіантів генів протеїну 3, що містить пататин-подібний фосфоліпазний домен, і трансмембранного протеїну 6 — члена суперродини 2, ймовірно, обумовлений посиленням експресії генів, що беруть участь у ліпогенезі de novo. Автори наголошують на необхідності оцінки генетичного ризику МАЖХП, що має включати молекулярно-генетичні дослідження на ранньому етапі обстеження.
The literature review highlights the issue of genetic risk factors associated with the development of metabolic dysfunction-associated fatty liver disease. Human genetic examinations revealed 132 genes among which 32 loci are strongly associated with the pathogenesis of metabolic dysfunction-associated fatty liver disease. It has been found that the risk of developing metabolic dysfunction-associated fatty liver disease is carried by single-nucleotide variants of various genes whose products are involved in lipid and carbohydrate metabolism, maintenance of the redox state, the development of inflammation and fibrosis of liver tissue, which are components of metabolic dysfunction-associated fatty liver disease reactome. The authors presented a detailed list of genetic factors singling out those that influence the risk of metabolic dysfunction-associated fatty liver disease and directly metabolic dysfunction-associated steatohepatitis and liver fibrosis. Also, they emphasized that it is the single-nucleotide variants of the genes of protein 3 containing a patatin-like phospholipase domain, transmembrane 6 superfamily member 2, and 17b-hydroxysteroid dehydrogenase type 13 that are characterized by the highest degree of association with metabolic dysfunction-associated fatty liver disease (odds ratio > 1.6) compared to single-nucleotide variants of other genes identified by gene association studies. The combination of several polymorphisms increases the risk of development and severity of metabolic dysfunction-associated fatty liver disease. The additive steatogenic effect of protein 3 single-nucleotide gene variants containing a patatin-like phospholipase domain and transmembrane 6 superfamily member 2 is probably due to an increased expression of genes involved in de novo lipogenesis. The authors emphasize the need for genetic risk assessment of metabolic dysfunction-associated fatty liver disease, which should include molecular genetic testing at an early stage of examination.
метаболічно асоційована жирова хвороба печінки; генетичні фактори ризику; ожиріння; діти
metabolic dysfunction-associated fatty liver disease; genetic disposition; obesity; children
Для ознакомления с полным содержанием статьи необходимо оформить подписку на журнал.
- Cui J., Chen C.H., Lo M.T., Schork N., Bettencourt R., Gonzalez M.P., et al.; For The Genetics Of Nafld In Twins Consortium. Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology. 2016 Nov;64(5):1547-1558. doi: 10.1002/hep.28674.
- Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021 May 13;184(10):2537-2564. doi: 10.1016/j.cell.2021.04.015.
- Wajsbrot N.B., Leite N.C., Salles G.F., Villela-Nogueira C.A. Non-alcoholic fatty liver disease and the impact of genetic, epigenetic and environmental factors in the offspring. World J Gastroenterol. 2022 Jul 7;28(25):2890-2899. doi: 10.3748/wjg.v28.i25.2890.
- Caussy C., Soni M., Cui J., Bettencourt R., Schork N., Chen C.H., et al.; Familial NAFLD Cirrhosis Research Consortium. Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J Clin Invest. 2017 Jun 30;127(7):2697-2704.
- Tamaki N., Ahlholm N., Luukkonen P.K., Porthan K., Sharpton S.R., Ajmera V., et al. Risk of advanced fibrosis in first-degree relatives of patients with nonalcoholic fatty liver disease. J Clin Invest. 2022 Nov 1;132(21):e162513. doi: 10.1172/JCI162513.
- Sookoian S., Pirola C.J. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol. 2017 Mar;23(1):1-12. doi: 10.3350/cmh.2016.0109.
- Sookoian S., Pirola C.J., Valenti L., Davidson N.O. Genetic Pathways in Nonalcoholic Fatty Liver Disease: Insights From Systems Biology. Hepatology. 2020 Jul;72(1):330-346. doi: 10.1002/hep.31229.
- Tidwell J., Wu G.Y. Unique Genetic Features of Lean NAFLD: A Review of Mechanisms and Clinical Implications. J Clin Transl Hepatol. 2024 Jan 28;12(1):70-78. doi: 10.14218/JCTH.2023.00252.
- Lin Y.C., Wu C.C., Ni Y.H. New Perspectives on Genetic Prediction for Pediatric Metabolic Associated Fatty Liver Disease. Front Pediatr. 2020 Dec 9;8:603654. doi: 10.3389/fped.2020.603654.
- Sookoian S., Pirola C.J. Genetics in non-alcoholic fatty liver disease: The role of risk alleles through the lens of immune response. Clin Mol Hepatol. 2023 Feb;29(Suppl):S184-S195. doi: 10.3350/cmh.2022.0318.
- Idilman R., Karatayli S.C., Kabacam G., Savas B., Elhan A.H., Bozdayi A.M. The role of PNPLA3 (rs738409) C>G variant on histological progression of non-alcoholic fatty liver disease. Hepatol Forum. 2020 Sep 21;1(3):82-87. doi: 10.14744/hf.2020.2020.0023.
- Meroni M., Dongiovanni P. PNPLA3 rs738409 Genetic Variant Inversely Correlates with Platelet Count, Thereby Affecting the Performance of Noninvasive Scores of Hepatic Fibrosis. Int J Mol Sci. 2023 Oct 10;24(20):15046. doi: 10.3390/ijms242015046.
- Abaturov A., Nikulina A. Role of genetic modification of the PNPLA3 gene in predicting metabolically unhealthy obesity and associated fatty liver disease in children. Eur J Clin Exp Med. 2023;21(1):5-13. doi: 10.15584/ejcem.2023.1.1.
- Kawaguchi T., Shima T., Mizuno M., Mitsumoto Y., Umemura A., Kanbara Y., et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS One. 2018 Jan 31;13(1):e0185490. doi: 10.1371/journal.pone.0185490.
- Ma’ruf M., Irham L.M., Adikusuma W., Sarasmita M.A., Khairi S., Purwanto B.D., et al. A genomic and bioinformatic-based approach to identify genetic variants for liver cancer across multiple continents. Genomics Inform. 2023 Dec;21(4):e48. doi: 10.5808/gi.23067.
- Kubiliun M.J., Cohen J.C., Hobbs H.H., Kozlitina J. Contribution of a genetic risk score to ethnic differences in fatty liver disease. Liver Int. 2022 Oct;42(10):2227-2236. doi: 10.1111/liv.15322.
- Li X.Y., Liu Z., Li L. Wang H.J., Wang H. TM6SF2 rs58542926 is related to hepatic steatosis, fibrosis and serum lipids both in adults and children: A meta-analysis. Front Endocrinol (Lausanne). 2022 Oct 24;13:1026901. doi: 10.3389/fendo.2022.1026901.
- Pourteymour S., Drevon C.A., Dalen K.T., Norheim F.A. Mechanisms Behind NAFLD: a System Genetics Perspective. Curr Atheroscler Rep. 2023 Nov;25(11):869-878. doi: 10.1007/s11883-023-01158-3.
- Mohammadi S., Farajnia S., Shadmand M., Mohseni F., Baghban R. Association of rs780094 polymorphism of glucokinase regulatory protein with non-alcoholic fatty liver disease. BMC Res Notes. 2020 Jan 10;13(1):26. doi: 10.1186/s13104-020-4891-y.
- Yuan F., Gu Z., Bi Y., Yuan R., Niu W., Ren D., Zhang L., et al. The association between rs1260326 with the risk of NAFLD and the mediation effect of triglyceride on NAFLD in the elderly Chinese Han population. Aging (Albany NY). 2022 Mar 25;14(6):2736-2747. doi: 10.18632/aging.203970.
- Bellini G., Miraglia Del Giudice E., Nobili V., Rossi F. The IRGM rs10065172 variant increases the risk for steatosis but not for liver damage progression in Italian obese children. J Hepatol. 2017 Sep;67(3):653-655. doi: 10.1016/j.jhep.2017.02.037.
- Ismaiel A., Spinu M., Osan S., Leucuta D.C., Popa S.L., Chis B.A., Farcas M., et al. MBOAT7 rs641738 variant in metabolic-dysfunction-associated fatty liver disease and cardiovascular risk. Med Pharm Rep. 2023 Jan;96(1):41-51. doi: 10.15386/mpr-2504.
- Tang S., Zhang J., Mei T.T., Zhang W.Y., Zheng S.J., Yu H.B. Association of HSD17B13 rs72613567: TA allelic variant with liver disease: review and meta-analysis. BMC Gastroenterol. 2021 Dec 20;21(1):490. doi: 10.1186/s12876-021-02067-y.
- Ting Y.W., Kong A.S., Zain S.M., Chan W.K., Tan H.L., Mohamed Z., et al. Loss-of-function HSD17B13 variants, non-alcoholic steatohepatitis and adverse liver outcomes: Results from a multi-ethnic Asian cohort. Clin Mol Hepatol. 2021 Jul;27(3):486-498. doi: 10.3350/cmh.2020.0162.
- Yoo T., Joo S.K., Kim H.J., Kim H.Y., Sim H., Lee J., Kim H.H., et al.; Innovative Target Exploration of NAFLD (ITEN) consortium. Disease-specific eQTL screening reveals an anti-fibrotic effect of AGXT2 in non-alcoholic fatty liver disease. J Hepatol. 2021 Sep;75(3):514-523. doi: 10.1016/j.jhep.2021.04.011.
- Wang J., Ye C., Fei S. Association between APOC3 polymorphisms and non-alcoholic fatty liver disease risk: a meta-analysis. Afr Health Sci. 2020 Dec;20(4):1800-1808. doi: 10.4314/ahs.v20i4.34.
- Zain S.M., Mohamed Z., Mahadeva S., Rampal S., Basu R.C., Cheah P.L., et al. Susceptibility and gene interaction study of the angiotensin II type 1 receptor (AGTR1) gene polymorphisms with non-alcoholic fatty liver disease in a multi-ethnic population. PLoS One. 2013;8(3):e58538. doi: 10.1371/journal.pone.0058538.
- Chaaba R., Bouaziz A., Ben Amor A., Mnif W., Hammami M., Mehri S. Fatty Acid Profile and Genetic Variants of Proteins Involved in Fatty Acid Metabolism Could Be Considered as Disease Predictor. Diagnostics (Basel). 2023 Mar 4;13(5):979. doi: 10.3390/diagnostics13050979.
- Mu T., Peng L., Xie X., He H., Shao Q., Wang X., Zhang Y. Single Nucleotide Polymorphism of Genes Associated with Metabolic Fatty Liver Disease. J Oncol. 2022 Feb 3;2022:9282557. doi: 10.1155/2022/9282557.
- Severson T.J., Besur S., Bonkovsky H.L. Genetic factors that affect nonalcoholic fatty liver disease: A systematic clinical review. World J Gastroenterol. 2016 Aug 7;22(29):6742-56. doi: 10.3748/wjg.v22.i29.6742.
- Ji F., Liu Y., Hao J.G., Wang L.P., Dai M.J., Shen G.F., Yan X.B. KLB gene polymorphism is associated with obesity and non-alcoholic fatty liver disease in the Han Chinese. Aging (Albany NY). 2019 Sep 23;11(18):7847-7858. doi: 10.18632/aging.102293.
- Valenti L., Motta B.M., Alisi A., Sartorelli R., Buonaiuto G., Dongiovanni P., Rametta R., et al. LPIN1 rs13412852 polymorphism in pediatric nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2012 May;54(5):588-93. doi: 10.1097/MPG.0b013e3182442a55.
- Yuan C., Lu L., An B., Jin W., Dong Q., Xin Y., Xuan S. Association Between LYPLAL1 rs12137855 Polymorphism With Ultrasound-Defined Non-Alcoholic Fatty Liver Disease in a Chinese Han Population. Hepat Mon. 2015 Dec 19;15(12):e33155. doi: 10.5812/hepatmon.33155.
- Luukkonen P.K., Juuti A., Sammalkorpi H., Penttilä A.K., Orešič M., Hyötyläinen T., Arola J., et al. MARC1 variant rs2642438 increases hepatic phosphatidylcholines and decreases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2020 Sep;73(3):725-726. doi: 10.1016/j.jhep.2020.04.021.
- Dai D., Wen F., Zhou S., Su Z., Liu G., Wang M, Zhou J., et al. Association of MTTP gene variants with pediatric NAFLD: A candidate-gene-based analysis of single nucleotide variations in obese children. PLoS One. 2017 Sep 27;12(9):e0185396. doi: 10.1371/journal.pone.0185396.
- Xu K., Zheng K.I., Zhu P.W., Liu W.Y., Ma H.L., Li G., Tang L.J., et al. Interaction of SAMM50-rs738491, PARVB-rs5764455 and PNPLA3-rs738409 Increases Susceptibility to Nonalcoholic Steatohepatitis. J Clin Transl Hepatol. 2022 Apr 28;10(2):219-229. doi: 10.14218/JCTH.2021.00067.
- Piras I.S., Raju A., Don J., Schork N.J., Gerhard G.S., DiStefano J.K. Hepatic PEMT Expression Decreases with Increasing NAFLD Severity. Int J Mol Sci. 2022 Aug 18;23(16):9296. doi: 10.3390/ijms23169296.
- Zhang R.N., Shen F., Pan Q., Cao H.X., Chen G.Y., Fan J.G. PPARGC1A rs8192678 G>A polymorphism affects the severity of hepatic histological features and nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease. World J Gastroenterol. 2021 Jul 7;27(25):3863-3876. doi: 10.3748/wjg.v27.i25.3863.
- Yoneda M., Hotta K., Nozaki Y., Endo H., Uchiyama T., Mawatari H., et al. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD). BMC Gastroenterol. 2008 Jun 27;8:27. doi: 10.1186/1471-230X-8-27.
- Hernaez R., McLean J., Lazo M., Brancati F.L., Hirschhorn J.N., Borecki I.B., et al.; Genetics of Obesity-Related Liver Disease (GOLD) Consortium; Nguyen T., Kamel I.R., Bonekamp S., Eber–hardt M.S., Clark J.M., Kao W.H., et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013 Sep;11(9):1183-1190.e2. doi: 10.1016/j.cgh.2013.02.011.
- Di Costanzo A., Belardinilli F., Bailetti D., Sponziello M., D’Erasmo L., Polimeni L., et al. Evaluation of Polygenic Determinants of Non-Alcoholic Fatty Liver Disease (NAFLD) By a Candidate Genes Resequencing Strategy. Sci Rep. 2018 Feb 27;8(1):3702. doi: 10.1038/s41598-018-21939-0.
- Abshagen K., Berger C., Dietrich A., Schütz T., Wittekind C., Stumvoll M., et al. A Human REPIN1 Gene Variant: Genetic Risk Factor for the Development of Nonalcoholic Fatty Liver Disease. Clin Transl Gastroenterol. 2020 Jan;11(1):e00114. doi: 10.14309/ctg.0000000000000114.
- Zhao J., Xu X., Wei X., Zhang S., Xu H., Wei X., et al. SAMM50-rs2073082, -rs738491 and -rs3761472 Interactions Enhancement of Susceptibility to Non-Alcoholic Fatty Liver Disease. Biomedicines. 2023 Aug 29;11(9):2416. doi: 10.3390/biomedicines11092416.
- Qiao M., Yang J.H., Zhu Y., Hu J.P. Association of sorting and assembly machinery component 50 homolog gene polymorphisms with nonalcoholic fatty liver disease susceptibility. Medicine (Baltimore). 2022 Jul 22;101(29):e29958. doi: 10.1097/MD.0000000000029958.
- Wu N., Zhai X., Yuan F., Li J., Yu N., Zhang F., Li D., et al. Fasting glucose mediates the influence of genetic variants of SOD2 gene on lean non-alcoholic fatty liver disease. Front Genet. 2022 Oct 18;13:970854. doi: 10.3389/fgene.2022.970854.
- Liu Q., Liu S.S., Zhao Z.Z., Zhao B.T., Du S.X., Jin W.W., Xin Y.N. TRIB1 rs17321515 gene polymorphism increases the risk of coronary heart disease in general population and non-alcoholic fatty liver disease patients in Chinese Han population. Lipids Health Dis. 2019 Aug 31;18(1):165. doi: 10.1186/s12944-019-1108-2.
- Xu Y.P., Liang L., Wang C.L., Fu J.F., Liu P.N., Lv L.Q., Zhu Y.M. Association between UCP3 gene polymorphisms and nonalcoholic fatty liver disease in Chinese children. World J Gastroenterol. 2013 Sep 21;19(35):5897-903. doi: 10.3748/wjg.v19.i35.5897.
- Zhang P.P., Song J.Y., Wang H.J., Wang H. [Combined effect of PNPLA3 rs738409 and UGT1A1 rs10929303 gene polymorphisms on nonalcoholic fatty liver disease in children]. Zhonghua Gan Zang Bing Za Zhi. 2023 Jul 20;31(7):723-728. Chinese. doi: 10.3760/cma.j.cn501113-20220222-00084.
- Zusi C., Mantovani A., Olivieri F., Morandi A., Corradi M., Miraglia Del Giudice E., et al. Contribution of a genetic risk score to clinical prediction of hepatic steatosis in obese children and adolescents. Dig Liver Dis. 2019 Nov;51(11):1586-1592. doi: 10.1016/j.dld.2019.05.029.
- Kozlitina J. Genetic Risk Factors and Disease Modifiers of Nonalcoholic Steatohepatitis. Gastroenterol Clin North Am. 2020 Mar;49(1):25-44. doi: 10.1016/j.gtc.2019.09.001.
- Park H., Yoon E.L., Chung G.E., Choe E.K., Bae J.H., Choi S.H., et al. Genetic and Metabolic Characteristics of Lean Nonalcoholic Fatty Liver Disease in a Korean Health Examinee Cohort. Gut Liver. 2024 Mar 15;18(2):316-327. doi: 10.5009/gnl230044.
- Tidwell J., Wu G.Y. Unique Genetic Features of Lean NAFLD: A Review of Mechanisms and Clinical Implications. J Clin Transl Hepatol. 2024 Jan 28;12(1):70-78. doi: 10.14218/JCTH.2023.00252.
- Pingitore P., Romeo S. The role of PNPLA3 in health and disease. Biochim Biophys Acta Mol Cell Biol Lipids. 2019 Jun;1864(6):900-906. doi: 10.1016/j.bbalip.2018.06.018.
- Lulić A.M., Katalinić M. The PNPLA family of enzymes: characterisation and biological role. Arh Hig Rada Toksikol. 2023 Jun 26;74(2):75-89. doi: 10.2478/aiht-2023-74-3723.
- Tavaglione F., Targher G., Valenti L., Romeo S. Human and molecular genetics shed lights on fatty liver disease and diabetes conundrum. Endocrinol Diabetes Metab. 2020 Sep 4;3(4):e00179. doi: 10.1002/edm2.179.
- Dhar D., Loomba R. Emerging Metabolic and Transcriptomic Signature of PNPLA3-Associated NASH. Hepatology. 2021 Apr;73(4):1248-1250. doi: 10.1002/hep.31735.
- Sherman D.J., Liu L., Mamrosh J.L., Xie J., Ferbas J., Lomenick B., et al. The fatty liver disease-causing protein PNPLA3-I148M alters lipid droplet-Golgi dynamics. bioRxiv [Preprint]. 2023 Oct 14:2023.10.13.562302. doi: 10.1101/2023.10.13.562302.
- Xiang H., Wu Z., Wang J., Wu T. Research progress, challenges and perspectives on PNPLA3 and its variants in Liver Diseases. J Cancer. 2021 Aug 13;12(19):5929-5937. doi: 10.7150/jca.57951.
- Banini B.A., Kumar D.P., Cazanave S., Seneshaw M., Mirshahi F., Santhekadur P.K., et al. Identification of a Metabolic, Transcriptomic, and Molecular Signature of Patatin-Like Phospholipase Domain Containing 3-Mediated Acceleration of Steatohepatitis. Hepatology. 2021 Apr;73(4):1290-1306. doi: 10.1002/hep.31609.
- Park J., Zhao Y., Zhang F., Zhang S., Kwong A.C., Zhang Y., et al. IL-6/STAT3 axis dictates the PNPLA3-mediated susceptibility to non-alcoholic fatty liver disease. J Hepatol. 2023 Jan;78(1):45-56. doi: 10.1016/j.jhep.2022.08.022.
- Wagner C., Hois V., Pajed L., Pusch L.M., Wolinski H., Trau–ner M., et al. Lysosomal acid lipase is the major acid retinyl ester hydrolase in cultured human hepatic stellate cells but not essential for retinyl ester degradation. Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Aug;1865(8):158730. doi: 10.1016/j.bbalip.2020.158730.
- Pirazzi C., Valenti L., Motta B.M., Pingitore P., Hedfalk K., Mancina R.M., et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet. 2014 Aug 1;23(15):4077-85. doi: 10.1093/hmg/ddu121.
- Bruschi F.V., Tardelli M., Einwallner E., Claudel T., Trau–ner M. PNPLA3 I148M Up-Regulates Hedgehog and Yap Signaling in Human Hepatic Stellate Cells. Int J Mol Sci. 2020 Nov 18;21(22):8711. doi: 10.3390/ijms21228711.
- Gou Y., Wang L., Zhao J., Xu X., Xu H., Xie F., et al. PNPLA3-I148M Variant Promotes the Progression of Liver Fibrosis by Inducing Mitochondrial Dysfunction. Int J Mol Sci. 2023 Jun 2;24(11):9681. doi: 10.3390/ijms24119681.
- Rady B., Nishio T., Dhar D., Liu X., Erion M., Kisseleva T., et al. PNPLA3 downregulation exacerbates the fibrotic response in human hepatic stellate cells. PLoS One. 2021 Dec 8;16(12):e0260721. doi: 10.1371/journal.pone.0260721.
- Salari N., Darvishi N., Mansouri K., Ghasemi H., Hosseinian-Far M., Darvishi F., Mohammadi M. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease: a systematic review and meta-analysis. BMC Endocr Disord. 2021 Jun 19;21(1):125. doi: 10.1186/s12902-021-00789-4.
- Shi F., Zhao M., Zheng S., Zheng L., Wang H. Advances in genetic variation in metabolism-related fatty liver disease. Front Genet. 2023 Sep 11;14:1213916. doi: 10.3389/fgene.2023.1213916.
- Li Y., Liu S., Gao Y., Ma H., Zhan S., Yang Y., et al. Association of TM6SF2 rs58542926 gene polymorphism with the risk of non-alcoholic fatty liver disease and colorectal adenoma in Chinese Han population. BMC Biochem. 2019 Feb 6;20(1):3. doi: 10.1186/s12858-019-0106-3.
- Kozlitina J., Smagris E., Stender S., Nordestgaard B.G., Zhou H.H., Tybjærg-Hansen A., et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014 Apr;46(4):352-6. doi: 10.1038/ng.2901.
- Maier S., Wieland A., Cree-Green M., Nadeau K., Sullivan S., Lanaspa M.A., et al. Lean NAFLD: an underrecognized and challenging disorder in medicine. Rev Endocr Metab Disord. 2021 Jun;22(2):351-366. doi: 10.1007/s11154-020-09621-1.
- Luo F., Smagris E., Martin S.A., Vale G., McDonald J.G., Fletcher J.A., et al. Hepatic TM6SF2 Is Required for Lipidation of VLDL in a Pre-Golgi Compartment in Mice and Rats. Cell Mol Gastroenterol Hepatol. 2022;13(3):879-899. doi: 10.1016/j.jcmgh.2021.12.008.
- Hussain M.M. Intestinal lipid absorption and lipoprotein formation. Curr Opin Lipidol. 2014 Jun;25(3):200-6. doi: 10.1097/MOL.0000000000000084.
- Reyes-Soffer G., Liu J., Thomas T., Matveyenko A., Seid H., Ramakrishnan R., et al. TM6SF2 Determines Both the Degree of Lipidation and the Number of VLDL Particles Secreted by the Liver. medRxiv [Preprint]. 2023 Jun 29:2023.06.23.23291823. doi: 10.1101/2023.06.23.23291823.
- Liu J., Ginsberg H.N., Reyes-Soffer G. Basic and translational evidence supporting the role of TM6SF2 in VLDL metabolism. Curr Opin Lipidol. 2024 Mar 11. doi: 10.1097/MOL.0000000000000930.
- Jiang X., Qian H., Ding W.X. New Glance at the Role of TM6SF2 in Lipid Metabolism and Liver Cancer. Hepatology. 2021 Sep;74(3):1141-1144. doi: 10.1002/hep.31851.
- Borén J., Adiels M., Björnson E., Matikainen N., Söderlund S., Rämö J., et al. Effects of TM6SF2 E167K on hepatic lipid and very low-density lipoprotein metabolism in humans. JCI Insight. 2020 Dec 17;5(24):e144079. doi: 10.1172/jci.insight.144079.
- Newberry E.P., Hall Z., Xie Y., Molitor E.A., Bayguinov P.O., Strout G.W., et al. Liver-Specific Deletion of Mouse Tm6sf2 Promotes Steatosis, Fibrosis, and Hepatocellular Cancer. Hepatology. 2021 Sep;74(3):1203-1219. doi: 10.1002/hep.31771.
- Trépo E., Valenti L. Update on NAFLD genetics: From new variants to the clinic. J Hepatol. 2020 Jun;72(6):1196-1209. doi: 10.1016/j.jhep.2020.02.020.
- Xue W.Y., Zhang L., Liu C.M., Gao Y., Li S.J., Huai Z.Y., et al. Research progress on the relationship between TM6SF2 rs58542926 polymorphism and non-alcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2022 Feb;16(2):97-107. doi: 10.1080/17474124.2022.2032661.
- Buchynskyi M., Oksenych V., Kamyshna I., Vari S.G., Kamyshnyi A. Genetic Predictors of Comorbid Course of COVID-19 and MAFLD: A Comprehensive Analysis. Viruses. 2023 Aug 12;15(8):1724. doi: 10.3390/v15081724.
- Luo F., Oldoni F., Das A. TM6SF2: A Novel Genetic Player in Nonalcoholic Fatty Liver and Cardiovascular Disease. Hepatol Commun. 2022 Mar;6(3):448-460. doi: 10.1002/hep4.1822.
- Sulaiman S.A., Dorairaj V., Adrus M.N.H. Genetic Polymorphisms and Diversity in Nonalcoholic Fatty Liver Disease (NAFLD): A Mini Review. Biomedicines. 2022 Dec 30;11(1):106. doi: 10.3390/biomedicines11010106.
- Meroni M., Longo M., Rustichelli A., Dongiovanni P. Nutrition and Genetics in NAFLD: The Perfect Binomium. Int J Mol Sci. 2020 Apr 23;21(8):2986. doi: 10.3390/ijms21082986.
- Schwerbel K., Kamitz A., Krahmer N., Hallahan N., Jähnert M., Gottmann P., et al. Immunity-related GTPase induces lipophagy to prevent excess hepatic lipid accumulation. J Hepatol. 2020 Oct;73(4):771-782. doi: 10.1016/j.jhep.2020.04.031.
- Xia M., Zeng H., Wang S., Tang H., Gao X. Insights into contribution of genetic variants towards the susceptibility of MAFLD revealed by the NMR-based lipoprotein profiling. J Hepatol. 2021 Apr;74(4):974-977. doi: 10.1016/j.jhep.2020.10.019.
- Matschinsky F.M., Wilson D.F. The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front Physiol. 2019 Mar 6;10:148. doi: 10.3389/fphys.2019.00148.
- Raimondo A., Rees M.G., Gloyn A.L. Glucokinase regulatory protein: complexity at the crossroads of triglyceride and glucose metabolism. Curr Opin Lipidol. 2015 Apr;26(2):88-95. doi: 10.1097/MOL.0000000000000155.
- Bianco C., Casirati E., Malvestiti F., Valenti L. Genetic predisposition similarities between NASH and ASH: Identification of new therapeutic targets. JHEP Rep. 2021 Mar 30;3(3):100284. doi: 10.1016/j.jhepr.2021.100284.
- Zhang Z., Ji G., Li M. Glucokinase regulatory protein: a balancing act between glucose and lipid metabolism in NAFLD. Front Endocrinol (Lausanne). 2023 Aug 29;14:1247611. doi: 10.3389/fendo.2023.1247611.
- Carlsson B., Lindén D., Brolén G., Liljeblad M., Bjursell M., Romeo S., Loomba R. Review article: the emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2020 Jun;51(12):1305-1320. doi: 10.1111/apt.15738.
- Ford B.E., Chachra S.S., Rodgers K., Moonira T., Al-Oanzi Z.H., Anstee Q.M., et al. The GCKR-P446L gene variant predisposes to raised blood cholesterol and lower blood glucose in the P446L mouse — a model for GCKR rs1260326. Mol Metab. 2023 Jun;72:101722. doi: 10.1016/j.molmet.2023.101722.
- Kang H., You H.J., Lee G., Lee S.H., Yoo T., Choi M., et al.; Innovative Target Exploration of NAFLD (ITEN) consortium. Interaction effect between NAFLD severity and high carbohydrate diet on gut microbiome alteration and hepatic de novo lipogenesis. Gut Microbes. 2022 Jan-Dec;14(1):2078612. doi: 10.1080/19490976.2022.2078612.
- Pusec C.M., Ilievski V., De Jesus A., Farooq Z., Zapater J.L., Sweis N., et al. Liver-specific overexpression of HKDC1 increases hepatocyte size and proliferative capacity. Sci Rep. 2023 May 17;13(1):8034. doi: 10.1038/s41598-023-33924-3.
- Iizuka K., Takao K., Yabe D. ChREBP-Mediated Regulation of Lipid Metabolism: Involvement of the Gut Microbiota, Liver, and Adipose Tissue. Front Endocrinol (Lausanne). 2020 Dec 3;11:587189. doi: 10.3389/fendo.2020.587189.
- Régnier M., Carbinatti T., Parlati L., Benhamed F., Postic C. The role of ChREBP in carbohydrate sensing and NAFLD development. Nat Rev Endocrinol. 2023 Jun;19(6):336-349. doi: 10.1038/s41574-023-00809-4.
- Iizuka K. The Roles of Carbohydrate Response Element Binding Protein in the Relationship between Carbohydrate Intake and Diseases. Int J Mol Sci. 2021 Nov 8;22(21):12058. doi: 10.3390/ijms222112058.
- Li J., Zhao Y., Zhang H., Hua W., Jiao W., Du X., et al. Contribution of Rs780094 and Rs1260326 Polymorphisms in GCKR Gene to Non-alcoholic Fatty Liver Disease: A Meta-Analysis Involving 26,552 Participants. Endocr Metab Immune Disord Drug Targets. 2021;21(9):1696-1708. doi: 10.2174/1871530320999201126202706.
- Nisar T., Arshad K., Abbas Z., Khan M.A., Safdar S., –Shaikh R.S., Saeed A. Prevalence of GCKR rs1260326 Variant in Subjects with Obesity Associated NAFLD and T2DM: A Case-Control Study in South Punjab, Pakistan. J Obes. 2023 Oct 4;2023:6661858. doi: 10.1155/2023/6661858.
- Wu N., Li J., Zhang J., Yuan F., Yu N., Zhang F., Li D., et al. Waist circumference mediates the association between rs1260326 in GCKR gene and the odds of lean NAFLD. Sci Rep. 2023 Apr 20;13(1):6488. doi: 10.1038/s41598-023-33753-4.
- Santoro N., Zhang C.K., Zhao H., Pakstis A.J., Kim G., Kursawe R., Dykas D.J., et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012 Mar;55(3):781-9. doi: 10.1002/hep.24806.
- Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., et al.; NASH CRN; GIANT Consortium; MAGIC Investigators; Voight B.F., Carr J.J., Feitosa M.F., Harris T.B., Fox C.S., Smith A.V., et al.; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011 Mar;7(3):e1001324. doi: 10.1371/journal.pgen.1001324.
- Anstee Q.M., Darlay R., Cockell S., Meroni M., Govaere O., Tiniakos D., et al.; EPoS Consortium Investigators. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol. 2020 Sep;73(3):505-515. doi: 10.1016/j.jhep.2020.04.003.
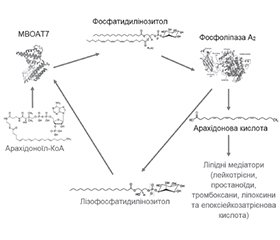
- Varadharajan V., Massey W.J., Brown J.M. Membrane-bound O-acyltransferase 7 (MBOAT7)-driven phosphatidylinositol remodeling in advanced liver disease. J Lipid Res. 2022 Jul;63(7):100234. doi: 10.1016/j.jlr.2022.100234.
- O’Donnell V.B. New appreciation for an old pathway: the Lands Cycle moves into new arenas in health and disease. Biochem Soc Trans. 2022 Feb 28;50(1):1-11. doi: 10.1042/BST20210579.
- Caddeo A., Spagnuolo R., Maurotti S. MBOAT7 in liver and extrahepatic diseases. Liver Int. 2023 Nov;43(11):2351-2364. doi: 10.1111/liv.15706.
- Massey W.J., Varadharajan V., Banerjee R., Brown A.L., Horak A.J., Hohe R.C., et al. MBOAT7-driven lysophosphatidylinositol acylation in adipocytes contributes to systemic glucose homeostasis. J Lipid Res. 2023 Apr;64(4):100349. doi: 10.1016/j.jlr.2023.100349.
- Meroni M., Longo M., Fracanzani A.L., Dongiovanni P. MBOAT7 down-regulation by genetic and environmental factors predisposes to MAFLD. EBioMedicine. 2020 Jul;57:102866. doi: 10.1016/j.ebiom.2020.102866.
- Thangapandi V.R., Knittelfelder O., Brosch M., Patsenker E., Vvedenskaya O., Buch S., et al. Loss of hepatic Mboat7 leads to liver fibrosis. Gut. 2021 May;70(5):940-950. doi: 10.1136/gutjnl-2020-320853.
- Teo K., Abeysekera K.W.M., Adams L., Aigner E., Anstee Q.M., Banales J.M., et al. rs641738C>T near MBOAT7 is associated with li–ver fat, ALT and fibrosis in NAFLD: A meta-analysis. J Hepatol. 2021 Jan;74(1):20-30. doi: 10.1016/j.jhep.2020.08.027.
- Xu X., Xu H., Liu X., Zhang S., Cao Z., Qiu L., et al. MBOAT7 rs641738 (C>T) is associated with NAFLD progression in men and decreased ASCVD risk in elder Chinese population. Front Endocrinol (Lausanne). 2023 Jun 22;14:1199429. doi: 10.3389/fendo.2023.1199429.
- Pan G., Cavalli M., Wadelius C. Polymorphisms rs55710213 and rs56334587 regulate SCD1 expression by modulating HNF4A bin–ding. Biochim Biophys Acta Gene Regul Mech. 2021 Aug;1864(8):194724. doi: 10.1016/j.bbagrm.2021.194724.
- Wang P., Wu C.X., Li Y., Shen N. HSD17B13 rs72613567 protects against liver diseases and histological progression of nonalcoholic fatty liver disease: a systematic review and meta-analysis. Eur Rev Med Pharmacol Sci. 2020 Sep;24(17):8997-9007. doi: 10.26355/eurrev_202009_22842.
- Su W., Mao Z., Liu Y., Zhang X., Zhang W., Gustafsson J.A., Guan Y. Role of HSD17B13 in the liver physiology and pathophysio–logy. Mol Cell Endocrinol. 2019 Jun 1;489:119-125. doi: 10.1016/j.mce.2018.10.014.
- Zhang H.B., Su W., Xu H., Zhang X.Y., Guan Y.F. HSD17B13: A Potential Therapeutic Target for NAFLD. Front Mol Biosci. 2022 Jan 7;8:824776. doi: 10.3389/fmolb.2021.824776.
- Su W., Wang Y., Jia X., Wu W., Li L., Tian X., et al. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci USA. 2014 Aug 5;111(31):11437-42. doi: 10.1073/pnas.1410741111.
- Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P., et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N Engl J Med. 2018 Mar 22;378(12):1096-1106. doi: 10.1056/NEJMoa1712191.
- Motomura T., Amirneni S., Diaz-Aragon R., Faccioli L.A.P., Malizio M.R., Coard M.C., et al. Is HSD17B13 Genetic Variant a Protector for Liver Dysfunction? Future Perspective as a Potential Therapeutic Target. J Pers Med. 2021 Jun 30;11(7):619. doi: 10.3390/jpm11070619.
- Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S., et al. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018 Apr;283(4):356-370. doi: 10.1111/joim.12719.
- Ji F., Liu Y., Hao J.G., Wang L.P., Dai M.J., Shen G.F., Yan X.B. KLB gene polymorphism is associated with obesity and non-alcoholic fatty liver disease in the Han Chinese. Aging (Albany NY). 2019 Sep 23;11(18):7847-7858. doi: 10.18632/aging.102293.
- Marzuillo P., Miraglia del Giudice E., Santoro N. Pediatric fatty liver disease: role of ethnicity and genetics. World J Gastroenterol. 2014 Jun 21;20(23):7347-55. doi: 10.3748/wjg.v20.i23.7347.
- Vestmar M.A., Andersson E.A., Christensen R., Hauge M., Glümer C., Linneberg A., et al. Functional and genetic epidemiological characterisation of the FFAR4 (GPR120) p.R270H variant in the Danish population. J Med Genet. 2016 Sep;53(9):616-23. doi: 10.1136/jmedgenet-2015-103728.
- Codoñer-Alejos A., Carrasco-Luna J., Codoñer-Franch P. The rs11187533 C>T Variant of the FFAR4 Gene Is Associated with Lower Levels of Fasting Glucose and Decreases in Markers of Liver Injury in Children with Obesity. Ann Nutr Metab. 2020;76(2):122-128. doi: 10.1159/000506618.
- Aaldijk A.S., Verzijl C.R.C., Jonker J.W., Struik D. Biological and pharmacological functions of the FGF19- and FGF21-coreceptor beta klotho. Front Endocrinol (Lausanne). 2023 May 16;14:1150222. doi: 10.3389/fendo.2023.1150222.
- Dongiovanni P., Crudele A., Panera N., Romito I., Meroni M., De Stefanis C., et al. β-Klotho gene variation is associated with liver damage in children with NAFLD. J Hepatol. 2020 Mar;72(3):411-419. doi: 10.1016/j.jhep.2019.10.011.
- Petta S., Valenti L., Marra F., Grimaudo S., Tripodo C., Bugianesi E., et al. MERTK rs4374383 polymorphism affects the severity of fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2016 Mar;64(3):682-90. doi: 10.1016/j.jhep.2015.10.016.
- Eslam M., Valenti L., Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol. 2018 Feb;68(2):268-279. doi: 10.1016/j.jhep.2017.09.003.
- Wang X., Cai B., Yang X., Sonubi O.O., Zheng Z., Ramakrishnan R., et al. Cholesterol Stabilizes TAZ in Hepatocytes to Promote Experimental Non-alcoholic Steatohepatitis. Cell Metab. 2020 May 5;31(5):969-986.e7. doi: 10.1016/j.cmet.2020.03.010.
- Al-Qarni R., Iqbal M., Al-Otaibi M., Al-Saif F., Alfadda A.A., Alkhalidi H., et al. Validating candidate biomarkers for different stages of non-alcoholic fatty liver disease. Medicine (Baltimore). 2020 Sep 4;99(36):e21463. doi: 10.1097/MD.0000000000021463.
- Canivet C.M., Bonnafous S., Rousseau D., Leclere P.S., La–cas-Gervais S., Patouraux S., et al. Hepatic FNDC5 is a potential local protective factor against Non-Alcoholic Fatty Liver. Biochim Biophys Acta Mol Basis Dis. 2020 May 1;1866(5):165705. doi: 10.1016/j.bbadis.2020.165705.
- Gao F., Zheng K.I., Wang X.B., Yan H.D., Sun Q.F., Pan K.H., et al. Metabolic associated fatty liver disease increases coronavirus disease 2019 disease severity in nondiabetic patients. J Gastroenterol Hepatol. 2021 Jan;36(1):204-207. doi: 10.1111/jgh.15112.
- Syafruddin S.E., Mohtar M.A., Wan Mohamad Nazarie W.F., Low T.Y. Two Sides of the Same Coin: The Roles of KLF6 in Physiology and Pathophysiology. Biomolecules. 2020 Sep 28;10(10):1378. doi: 10.3390/biom10101378.
- Holmer M., Ekstedt M., Nasr P., Zenlander R., Wester A., Tavaglione F., et al. Effect of common genetic variants on the risk of cirrhosis in non-alcoholic fatty liver disease during 20 years of follow-up. Liver Int. 2022 Dec;42(12):2769-2780. doi: 10.1111/liv.15438.
- Villani R., Magnati G.P., De Girolamo G., Sangineto M., Romano A.D., Cassano T., Serviddio G. Genetic Polymorphisms and Clinical Features in Diabetic Patients With Fatty Liver: Results From a Single-Center Experience in Southern Italy. Front Med (Lausanne). 2021 Oct 21;8:737759. doi: 10.3389/fmed.2021.737759.
- Paternostro R., Staufer K., Traussnigg S., Stättermayer A.F., Halilbasic E., Keritam O., et al. Combined effects of PNPLA3, TM6SF2 and HSD17B13 variants on severity of biopsy-proven non-alcoholic fatty liver disease. Hepatol Int. 2021 Aug;15(4):922-933. doi: 10.1007/s12072-021-10200-y.
- Ioannou G.N. Epidemiology and risk-stratification of NAFLD-associated HCC. J Hepatol. 2021 Dec;75(6):1476-1484. doi: 10.1016/j.jhep.2021.08.012.
- Jonas W., Schürmann A. Genetic and epigenetic factors determining NAFLD risk. Mol Metab. 2021 Aug;50:101111. doi: 10.1016/j.molmet.2020.101111.